Lab Notebook: Ecology of Marine Infectious Diseases
Sonia Singhal
22 Aug 2012
Labyrinthula lab:
Chasing after the (apparently non-specific) primers a bit...
- A BLAST search of both primers (GGAAGGCAGCAGGCGCGTAAnnnnnnnnnnnnnnnnCTATACCGACTGAGGATGGAAACATTC) turns up mostly Labyrinthula zosterae as top hits, followed by other ciliates, Desmarestia, and a few organisms that look like protists from the names. This does not explain why the primers are able to amplify bacterial DNA.
- However, the primers are located in small-subunit ribosomal sequences. These are highly conserved across taxa. Comparison of two labyrinthulids, even from opposite sides of the country (as Sammi Brombacker did) may not provide good enough resolution for the primers to work in practice.
- Ideal thing to do: Sequence some more specific regions of Labyrinthula (maybe microsatelites?) and align these among a couple Labyrinthula species and a closely related non-Labyrinthulid. Choosing some regions that are more variable across taxa may give better annealing resolution.
21 Aug 2012
Labyrinthula lab:
Discovered that the GoTaq Flexi mix is only a buffer-- that is, it contains no MgCl(2) or dNTPs. Lisa got us fresh stocks of each and also mentioned that the ImmoMix was unused by anyone in the class (although it required loading dye for visualization). Sammi and I decided to hedge our bets and run a PCR from each mix. We autoclaved the remainder of the PCR tubes in preparation. Reactions were set up under the Lab 10 hood, and tubes and water were UV irradiated beforehand.
Same reaction set used as 20 Aug.
PCR mix 1:
| Reagent |
µL for one reaction |
Total µL |
|
| 5x GoTaq flexi buffer |
4 |
X 21 |
84 |
| BSA |
1 |
21 |
|
| MgCl2 (25 uM) |
3 |
63 |
|
| dNTP (10 uM) |
2 |
42 |
|
| P1 |
0.5 |
10.5 |
|
| P2 |
0.5 |
10.5 |
|
| Taq |
0.2 |
4.2 |
|
| PCR water |
6.8 |
142.8 |
|
| Total |
18 |
378 |
PCR mix 2:
| Reagent |
µL for one reaction |
Total µL |
|
| ImmoMix |
12.5 |
X 21 |
262.5 |
| BSA |
1.5 |
31.5 |
|
| P1 |
0.8 |
16.8 |
|
| P2 |
0.8 |
16.8 |
|
| PCR water |
7.4 |
155.4 |
|
| Total |
23 |
483 |
Cycle (both mixes):
| Step |
Temperature (C) |
Time |
| 1 |
95 |
10 min |
| 40 cycles of: |
||
| 2 |
95 |
30 min |
| 3 |
50 |
30 sec |
| 4 |
72 |
60 sec |
| 5 |
72 |
10 min |
Visualization on a 2% agarose gel (60 min at 65V) showed no bands for the Flexi mix, but there were bands for the ImmoMix!
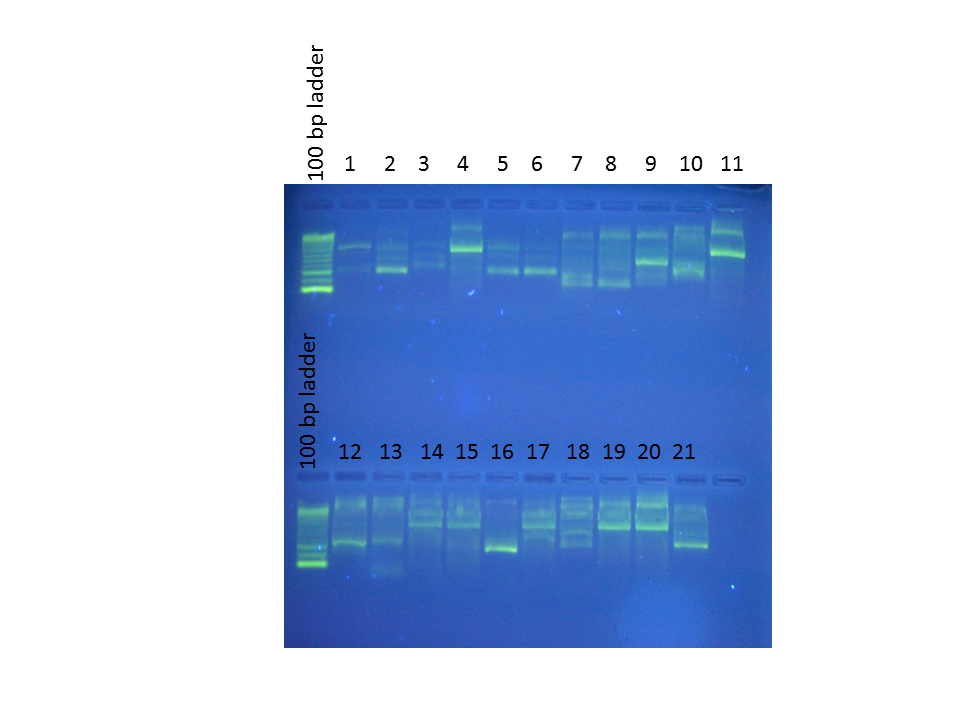
Order of wells:
1. Phylloplesia head
2. Phylloplesia gonads
3. Phylloplesia gizzard
4. Phylloplesia digestive tract
5. Phylloplesia nidamental gland
6. Phylloplesia posterior
7. Lacuna diseased 1
8. Lacuna diseased 2
9. Lacuna diseased 3
10. Lacuna healthy 1
11. Positive control
12. Lacuna healthy 2
13. Lacuna healthy 3
14. CK Shallow Bay culture
15. FB Sh T-1-D culture
16. FB Sh T-1 culture
17. Beach Haven (extraction 1 Aug)
18. Vibrio RE22
19. Sandy's monoculture
20. Positive control
21. Negative control
Results:
- Sammi got some good evidence that Laby is located in the Phylloplesia digestive tract, as opposed to being in other places.
- The Lacuna results are inconclusive.
- Cultures CK Shallow Bay, FB Sh-T-1-D, BH, and Monoculture seem to match the positive control.
- ...On the other hand, we also had amplification from Vibrio DNA, which means that our primers are not very specific.
- ...Also, we're amplifying something in the negative control in spite of all the care we took to stay sterile.
20 Aug 2012
Labyrinthula lab:
Visualization of PCR products from yesterday, run on 2% agarose gel for 60 min at 65 V (done because we're using the bad ladder that smeared last time). Photo not included because there were no bands whatsoever to be seen, including for the positive control. (The ladder, however, looked gorgeous.) We thought that perhaps since the power had gone out in the middle of the night, the PCR hadn't finished properly.
Re-ran the PCR. I added in previous BH and monoculture extractions.
PCR mix:
| Reagent |
µL for one reaction |
Total µL |
|
| 5x GoTaq flexi mix |
5 |
X 21 |
105 |
| BSA |
1.5 |
31.5 |
|
| P1 |
0.8 |
16.8 |
|
| P2 |
0.8 |
16.8 |
|
| PCR water |
14.65 |
307.65 |
|
| Taq |
0.25 |
5.25 |
|
| Total |
23 |
483 |
Forward, 5’ GGAAGGCAGCAGGCGCGTAA 3’
Reverse 5’ GAATGTTTCCATCCTCAGTCGGTATAG 3’
2 uL of each DNA sample added to each reaction. Positive control: Pool Laby + (stock); Negative control: PCR water
Cycle conditions:
| Step |
Temperature ( C) |
Time |
| 1 |
95 |
10 min |
| 40 cycles of: |
||
| 2 |
95 |
30 min |
| 3 |
50 |
30 sec |
| 4 |
72 |
60 sec |
| 5 |
72 |
10 min |
Reactions removed from the PCR machine as soon as they finished. Another gel visualization still showed... no bands whatsoever.
Maybe our sterile technique was too sterile?
19 Aug 2012
Labyrinthula lab:
Redo of DNA extraction and PCR:
Sammi and I autoclaved all tubes we wanted to use (1.5-mL, 2-mL, PCR) and got new aliquots of all materials. Lisa gave us an untouched GoTaq Flexi mix to use for PCRs.
Extractions as per 18 Aug-- only CK and FB cultures redone. Sammi dissected different parts of a Phylloplesia to try to isolate which organs Laby might be present in. I added a Vibrio (RE22) extraction as a definite negative control and did it completely under the hood--not as a sterile space, but as a Laby-free space. PCR tubes and water UV'ed under the hood for 20 min before use.
PCR mix: Flexi Taq
| Reagent |
µL for one reaction |
Total µL |
|
| 5x GoTaq flexi mix |
5 |
X 19 |
95 |
| BSA |
1.5 |
28.5 |
|
| P1 |
0.8 |
15.2 |
|
| P2 |
0.8 |
15.2 |
|
| PCR water |
14.65 |
278.35 |
|
| Taq |
0.25 |
4.75 |
|
| Total |
23 |
437 |
Forward, 5’ GGAAGGCAGCAGGCGCGTAA 3’
Reverse 5’ GAATGTTTCCATCCTCAGTCGGTATAG 3’
2 uL of each DNA sample added to each reaction. Positive control: Pool Laby + (stock); Negative control: PCR water
Cycle conditions:
| Step |
Temperature ( C) |
Time |
| 1 |
95 |
10 min |
| 40 cycles of: |
||
| 2 |
95 |
30 min |
| 3 |
50 |
30 sec |
| 4 |
72 |
60 sec |
| 5 |
72 |
10 min |
| 6 |
10 |
overnight |
18 Aug 2012
Labyrinthula lab:
DNA extraction and PCR of Labyrinthula cultures:
Plated cultures used:
- 1 : Sandy 1.17.1 from the eelgrass infection plate, 8/7
- 2 : Shallow Bay 1 CK, 7/18
- 3 : Replate of FB Sh-T-1-D, plated 8/11
- 4 : FB Sh-T-1, replate for Herbivores 8/11
Scraping of each culture put into 700 uL ASL; DNA extracted using the QIAgen Stool kit (protocol under 26 July). Sample #2 had the lid pop off during the centrifugation at Step 13, but no liquid was lost. All other lids remained closed.
Sammi extracted DNA from her Lacuna snails.
PCR mix: GoTaq
| Reagent |
µL for one reaction |
Total µL |
|
| GoTaq mix |
12.5 |
X 14 |
175 |
| BSA |
1.5 |
21 |
|
| P1 |
0.8 |
11.2 |
|
| P2 |
0.8 |
11.2 |
|
| PCR water |
7.4 |
103.6 |
|
| Total |
23 |
322 |
Forward, 5’ GGAAGGCAGCAGGCGCGTAA 3’
Reverse 5’ GAATGTTTCCATCCTCAGTCGGTATAG 3’
2 uL of each DNA sample added to each reaction. Positive control: Pool Laby + (stock); Negative control: PCR water
Cycle conditions:
| Step |
Temperature ( C) |
Time |
| 1 |
95 |
10 min |
| 40 cycles of: |
||
| 2 |
95 |
30 min |
| 3 |
50 |
30 sec |
| 4 |
72 |
60 sec |
| 5 |
72 |
10 min |
Visualization on 1.5% agarose gel:
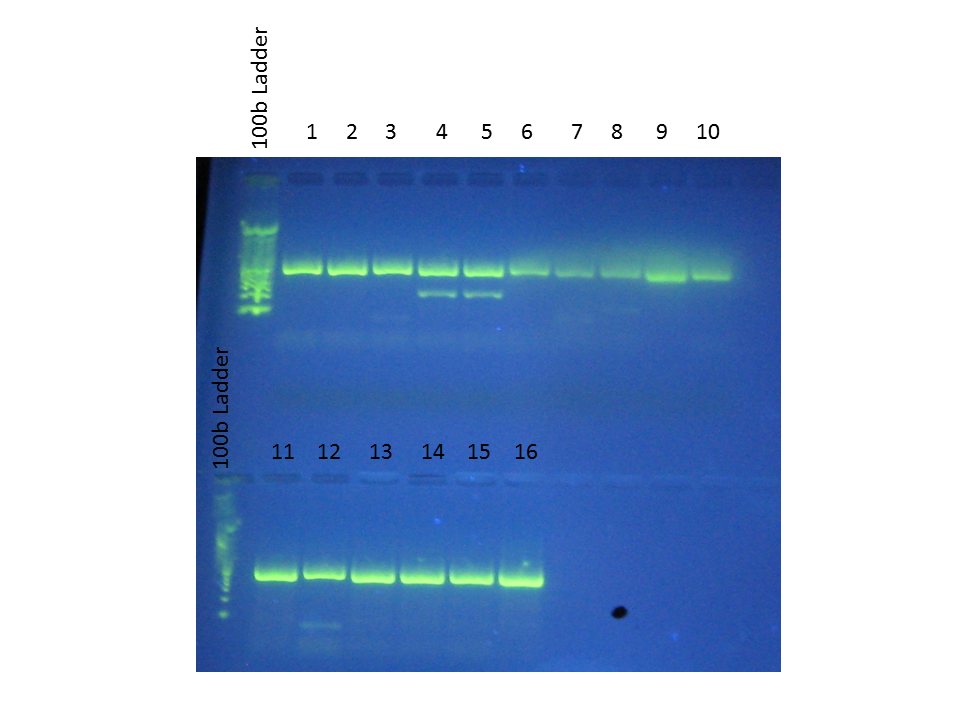
Order of wells:
1. Healthy Lacuna 1
2. Healthy Lacuna 2
3. Healthy Lacuna 3
4. Diseased Lacuna 1
5. Diseased Lacuna 1
6. Diseased Lacuna 2
7. Diseased Lacuna 3
8. * (previous Lacuna extraction?)
9. Positive control
10. Negative control
11. Laby culture 1
12. Laby culture 2
13. Laby culture 3
14. Laby culture 4
15. Beach Haven (extracted 1 Aug)
16. Positive control
...Because our negative control also shows bands, the results of the PCR are rather dubious. Since we were using the class reagents, something may have gotten contaminated...
17 Aug 2012
Bioinformatics lab:
- Started sifting through the table to find genes that overlap between GO-slim terms.
Labyrinthula lab:
Seawater took a long time to sterilize--autoclave kept aborting! Finally realized that it needed more water. Also got more bleach for sterilizing clips.
See 15 Aug for general protocol. Notes of details and modifications follow.
1. Culture inoculum
- Blades dipped in Betadine, then sterile seawater. Betadine worked extremely well--leaves still looked distinctively green afterward.
- Plates numbered separately for clip inocula and liquid inocula
- All blades photographed, cleaned and put in plates first
- Short mix-up with numbering, but it was cleared up.
- Scratches (3 small < 1cm) made longitudinally in about the middle of the blade with a pointed tool. Only broke the surface layers of cells
- Scratches accidentally put on ALL liquid inoculum blades. Because we had 7 blades left over, these were switched in for the scratched ones in some of the non-scratch plates: 3 control plates, 4 10^0 plates. Note, however, that these were blades that had initially been discarded.
- All other plates were filled as labeled. One plate was left over.
- Culture used: BH Laby culture used in the 10 Aug trial; 15 mL of the original (resuspended) culture had been kept going.
- Culture concentration: 8.6 x 10^6 => 3.44 x 10^4 cells/mL per plate in the 10^0 plates, 34.4 cells/mL per plate in the 10^(-2) plates.
- As before, the monoculture is too low a density to consider using.
2. Clip inoculum
The 1* culture from 6 Aug had grown over the inoculum leaves, so we additionally ran a second clip trial with that strain. Control plate looked uncontaminated, so 2 non-infected leaf-halves were included as negative controls.
Photos:
1*-1
1*-2
Control
Gregor and Courtney will be taking photos of all inoculated leaves ~ every 12 hours so that we can get time-series data on lesion development.
16 Aug 2012
Bioinformatics lab:
- Revised the scatterplot program so that it could create multiple pairwise graphs
Labyrinthula lab:
Setup for next inoculation experiments:
- Gathered supplies
- Cut 40 new blades—less particularity paid to the age of the blade/blade section, although I did my best to find the healthiest-looking pieces
- Autoclaved seawater
- Cut clips from fresh tubing
- Tested Betadine as an alternate sterilizing agent by dipping test leaves in a solution of ~ 10%. Leaves stayed green after dipping, in stark contrast to a set dipped in ethanol.
15 Aug 2012
Bioinformatics lab:
Check over- or under-representation of annotated genes between the full dataset and the differentially expressed data set.
- Use David: http://david.abcc.ncifcrf.gov/tools.jsp
- Create two text files, each with only one column. The first will have the Swiss Pro numbers of all the annotated genes. The other will have the Swiss Pro numbers of the differentially expressed genes.
- Click Start analysis on the top menu bar.
- In the left-hand dialogue box, Step 1, paste in the differentially expressed Swiss Pro list or upload it from the computer. Select “Uniprot_ID” from the drop-down menu in Step 2. Select “Gene list” in Step 3. Hit “Submit”.
- If the ID type was incorrect, submit the list to the Gene Conversion Tool. It will give a table with the proper ID type. At the top of the table, click the symbol that looks like pages and an arrow under “Convert all”. On the new page, click the green “Convert to Gene List” at the top of the table. David will then ask for a name.
- Repeat for the full SwissPro list, setting this one as “Background” in Step 3. Change to the proper ID type as well, if there were problems before.
- The resulting page will look roughly like this:
- Click on Functional Annotation Tool.
- Resulting page:
- GO has 3 sections: BP (biological), CC (cellular), and MP (molecular). The FAT file is a good middle-of-the-road. Click on “Chart” for more details.
- A new window will open containing a table. The table can be downloaded (upper right) for visualization on another program.
- We downloaded the BP_FAT table.
- In Excel, we only want the GO number. Choose Text to Column and change the tilde to a new column.
- Visualize: From the David spread, choose the GO number and p-value and paste them into the text box in Revigo (http://revigo.irb.hr/). It will give a page with a number of tabs containing interactive visualizations of the data.
- The data can also be downloaded as an R script for further modification. NOTE: Both the ggplot2 AND the scales library need to be installed for the script to work.
Labyrinthula lab:
Finalized the protocol for the reinfection experiments:
Protocol for Laby reinfection experiments
Approach 1: Blade inocula
Day 1: Preparation for plating
- Ensure that there are some plated Laby cultures growing that can be used as infectious agents.
- Cut healthy eelgrass blades into 1.5-3 cm pieces. Use the middle of the blades, avoiding any blemishes. Discard the very tips and the lighter green lower sections, which will fall apart after being autoclaved.
- Autoclave the blade cuttings in small amount of distilled water to sterilize them.
- Prepare plates with Laby media agar. Store at 4 C.
Day 2: Create the inocula
- Label all plates and sterilize scalpel and forceps before starting.
- WITH A FLAME GOING, open the growing Laby culture and cut a small piece of agar from a leading edge of the colony. Lever the cutting out right-side up with the scalpel and transfer to a clean plate. Turn the scalpel over and place the cutting on top of the clean agar, upside-down, without touching the scalpel to the clean agar.
- Using the forceps, add 2-4 blades to the petri dish. Sterilize (alcohol and flame) the forceps in between each blade.
- Cap the petri dish and parafilm.
- Repeat for all desired cultures for which you wish to obtain inocula.
- Add 1-2 control plates that receive autoclaved blades but no Laby.
- Incubate right-side up at 25-30 C.
Days 3-5: Laby platings will be growing over the agar and autoclaved leaves. This may take anywhere from 3 to 6 days, depending on how quickly the particular strain grows. When it looks like at least one blade has been surrounded by Laby, prepare for the inoculations:
- Cut healthy eelgrass leaves into pieces of ~ 8 cm and place in a tub of flowing seawater overnight (or longer) to allow the cuts to heal. Use pieces from the center of the leaf: discard 10-16 cm at each end. KEEP THE BLADES IN SEAWATER the entire time, because they can dry out rapidly in the air!
- Autoclave seawater to sterilize. You will need 25 mL for each blade you intend to inoculate.
- Create clips by cutting a length small, hollow tubing into pieces of approximately 1cm and slitting them lengthwise along one side. Sterilize by dipping them briefly in 10% bleach, then distilled water, a couple of times. Store overnight in fresh distilled water or sterilized seawater.
- Locate supplies:
- Empty petri dishes. Extras can be used to store instruments.
- Scalpel
- 2 forceps: one thinner pair and one thicker pair
- 70% ethanol
- Flame (Bunsen burner, lamp, etc.)
- 25-mL pipets and pipetman
- Betadine
- Ruler
- Camera
- Numbered paper towels or pieces of paper
- Create a randomized order for the inoculated blades. The blades can be inoculated in an order that makes sense for the cultures, but they will be photographed with their randomized number.
Day 6: Perform inoculations
- Keep a flame going whenever the inocula are being used.
- Label all petri dishes with the name and randomized number, and fill each with 25 mL of sterile seawater.
- Label a set of paper towels or paper squares with the randomized numbers.
- Set up camera in a stable position to take pictures. Each piece of eelgrass will be placed on the paper with its number and a cm ruler below it. If necessary, mark locations (of paper, ruler, camera tripod legs, etc.) on the table.
- Move the healed eelgrass to a bag of seawater. This does not have to be sterile.
- Transfer clips to a bath of sterile seawater.
- Workflow for healed blades. Use forceps.
- Remove blade from the bag of seawater. Place on numbered paper and photograph with a ruler.
- Dip blade briefly in Betadine, then wash in sterile seawater.
- Transfer blade to petri dish with the corresponding number.
- Workflow for inoculations:
- STERILIZE ALL INSTRUMENTS (ethanol, flame).
- The first person opens the petri dish with the autoclaved inoculum leaves. Use sterile forceps to remove one leaf.
- For 3-cm inoculum leaves, cut the leaf in half.
- The second person, with clean gloves, removes the healthy leaf from the petri dish and THEN takes the inoculum.
- The third person, with sterile wide-point forceps, removes a clip from the sterile seawater bath. The clip can be opened easily by putting the forceps into the tube and allowing them to open.
- The second person layers the inoculum on top of the healthy leaf and slips both into the opening of the clip. The leaf is replaced in the petri dish.
- Control blades receive inocula from the control agar plates.
- Sterilizations:
- The wide-point forceps are sterilized after each clip.
- The forceps used to retrieve inocula are sterilized before being put into ANY petri dish, even the same one.
- Scissors are sterilized before the next use, even if the inoculum has the same culture as the previous one.
- The person handling the inocula directly changes gloves after every inoculum.
- The person handling the agar plates changes gloves before handling a plate with a different inoculum.
- Randomize the petri dishes by placing them in numerical order. Incubate petri dishes at room temperature.
Day 7-10: Track lesion progress
Photograph petri dishes at 12-hour intervals until clear signs of lesions. Lesions will appear as dark streaks radiating from the center of the blade, where the inoculum was placed. Temperature data can also be taken.
Day 11: Analysis
Use ImageJ to analyze all blades. Protocol to be completed when Courtney goes over it with us.
Approach 2: Liquid culture inocula
Day 1-7: Start cultures
- Create Laby liquid cultures by taking an inoculation loop’s worth of an isolate from an agar plate and mixing it into 10 mL of Laby broth in 50-mL Falcon tubes.
- Incubate at room temperature.
- Note that Laby generally grows extremely slowly in broth, so prepare liquid cultures well ahead of the experiment.
Day 8: Setup for experiment
- Cut healthy eelgrass leaves into pieces of ~ 8 cm and place in a tub of flowing seawater overnight (or longer) to allow the cuts to heal. Use pieces from the center of the leaf: discard 10-16 cm at each end. KEEP THE BLADES IN SEAWATER the entire time, because they can dry out rapidly in the air!
- Autoclave seawater to sterilize. You will need 25 mL for each blade you intend to inoculate.
- Locate supplies:
- Empty petri dishes. Extras can be used to store instruments.
- Scalpel
- Pair of forceps
- 70% ethanol
- Flame (Bunsen burner, lamp, etc.)
- 25-mL pipets and pipetman
- Smaller-volume (< 1mL) pipets and tips
- Betadine
- Ruler
- Camera
- Numbered paper towels or pieces of paper
- Centrifuge for 50-mL Falcon tubes
- Hemocytometer
- Create a randomized order for the inoculated blades. The blades can be inoculated in an order that makes sense for the cultures, but they will be photographed with their randomized number.
Day 9: Inoculations, including scratch trial
- If multiple cultures of the same Laby isolate have been made, these can be pooled into a single Falcon tube and spun down (4000-6000 rpm for 15 min), then resuspended in 10-15 mL of fresh Laby broth.
- Cultures that have low concentrations of Laby can also be spun down and resuspended in a lower volume of broth.
- Mix cultures thoroughly before quantifying.
- Quantify cultures by hemocytometry. Use these numbers to determine the appropriate cultures and dilution factors to use.
- Label all petri dishes with the name and randomized number, and fill each with 25 mL of sterile seawater.
- Create a dilution series from the full culture (at least 100 µL of culture/ previous dilution in 900 µL sterile seawater).
- Mix dilutions thoroughly before use.
- Workflow for inoculations
- Using forceps, remove blade from the bag of seawater. Place on numbered paper and photograph with a ruler.
- Dip blade briefly in Betadine, then wash in sterile seawater.
- Add mechanical stress: brush blade center with a small wire brush or knick lightly (also in the center) with a sterile razor.
- Transfer blade to petri dish with the corresponding number.
- Inoculate the seawater in the dish with 100 µL of the appropriate culture dilution. Mix by swirling the petri dish gently.
- Controls:
- No Laby: These blades will receive 100 µL of fresh Laby broth.
- No stress: These blades will receive the Laby inoculum but will not be scratched beforehand.
- No Laby or stress: These blades will receive 100 µL of fresh Laby broth and no mechanical stress.
- Randomize the plates by placing in numerical order. Incubate at room temperature.
Day 10-14: Monitor progress of lesion growth
Photograph petri dishes at 12-hour intervals until clear signs of lesions. Ideally, lesions will appear first in the area where the blade was scratched and then expand. Temperature data can also be taken.
Day 15: Analysis in ImageJ.
14 Aug 2012
Bioinformatics lab:
Joining tables in Galaxy:
Galaxy’s setup is:
- Tools on the left
- Files and dialogue boxes in the middle
- History (steps taken, numbered in reverse chronological order) on the right. Every step is recorded, and steps that change or create new data tables are saved in the box of that step as new tables.
- “Eye” allows you to view the result of that step
- “Pencil” allows you to edit attributes of the file
- “X” cancels the step or deletes the table
- If you click on the title in the box, it will give you a “save” button that you can use to download the data table.
Protocol:
- Log into Galaxy (https://main.g2.bx.psu.edu/)
- Upload file(s) to Galaxy:
- Left bar: Get data
- Upload file from your computer (here, data = the qpx mapped reads)
- Check upload: Click on the “eye” symbol in the green box with the file name on the right-hand side.
- Separate different identifiers in Column 3 (column 2 in non-headed version):
- Left bar: Text manipulations
- Convert delimiters to tab
- In the dialogue box, make sure “pipes” are being converted.
- Check: Click on the “eye” in the right-hand green box labeled “Convert on data”. Can rename file if desired by clicking the “pencil”.
- Import previously-established history of SwissPro accession numbers and gene names:
- Go to https://main.g2.bx.psu.edu/u/sr320/h/databases.
- Click on the “eye” under “SPID and description”
- Click “upload” (green circle with the cross)
- Join two tables:
- MAKE SURE THE ROWS IN THE COLUMNS TO JOIN ARE IDENTICAL. (That is, that elements in the desired column in one table EXACTLY match the rows in the desired column in the other table.)
- Left bar: Join, subtract and group
- Join two datasets
- Here: In the dialogue box join “Convert on data” (or equivalent), c4 (c3 for non-headed version); with “SPID and description”, c1.
- Keep all lines and fill unjoined rows.
- Check: Click on the “eye” next to the “Join” box on the right.
- Add protein functions:
- Import the “SPID and GO #” database from the above address.
- Join two data tables as above, merging the columns with the SwissPro ID numbers.
- Check result. GO data should be in column 24 (23 in unheaded data table)
- Import the “GO and GO_slim” database from the above address.
- Join the data tables by GO number.
- Check result.
13 Aug 2012
Labyrinthula lab:
Reinfections:
Both Sandy’s culture and the 2* field culture show clear signs of lesions. We photographed all blade-inoculum plates both with and without the clips. The blades themselves were placed back into the same petri dish in the same place on the table, in case we want them for histology.
Liquid-inoculum plates will be left for a little longer to see if any infections develop later. We know that the cultures grow even more slowly than the plates, and not very many cells were inoculated into each plate.
Bioinformatics lab:
Downloaded the SwissProt database onto the lab computers (CL_14 à Users à Shared à EIMD_blast à db) and unzipped it. The database was then created through the terminal:
%% cd /Applications/blast/bin
%% ./makeblastdb -in [input file in .fasta] -out [output file name, no .fasta] -dbtype prot
Input: /Users/Shared/EIMD_blast/db/uniprot_sprot.fasta
Output: /Users/Shared/EIMD_blast/db/uniprot_sprot
- Note that the Applications folder used is also under CL_14, NOT under fhl_guest.
- On a Mac, directory can be set by dragging the appropriate folder from Finder into the Terminal.
Practice BLAST on the QPX transcriptome data, just to make sure it worked. Only the uniprot database worked; the viral database that was already loaded did not seem to.
%% ./blastx -query [input file in .fasta] -db [uniprot_sprot, no .fasta]
- Query: /Users/Shared/EIMD_blast/query/QPX_transcriptome_v1.fasta
- Database (db): /Users/Shared/EIMD_blast/db/uniprot_sprot
Started a BLAST of the full transcriptome:
First opened file (/Users/Shared/EIMD_blast/query/QPX_transcriptome_v1.fasta) in TextWrangler. (Say “No” to any options that come up when TextWrangler opens.) To prevent misreading of the sequence names/sequences, we did a reformat:
- Find: 1 (space) Contig (space)
- Replace all with: 1_Contig_
- Resave file.
In Terminal:
%% ./blastx
-query [input file]
-db [database file, no .fasta]
-evalue 1e-20
-max_target_seqs 1
-outfmt 6
-out [outfile]
-num_threads 2
Notes:
*query used: /Users/Shared/EIMD_blast/query/QPX_transcriptome_v1.fasta
*db: /Users/Shared/EIMD_blast/db/uniprot_sprot
*out: /Users/Shared/EIMD_blast/out/qpx_transcr_swisspro.txt
*E-value of e^(-20) => only HIGHLY similar sequences will be selected
*max_target_seqs – only return the first (highest) hit for each contig
*outfmt 6 = tab-delimited
*num_threads = number of CPUs to use
Note that the command above should be written in a single line, with spaces instead of enters/tabs. I’ve written it out that way to make it easier to read all the arguments.
Let the program run on the terminal overnight. Final location of the resulting text file:
https://catalyst.uw.edu/sharespaces/download/17360/318727?html=1&url=https://catalyst.uw.edu/sharespaces/download/17360/318727
Headed text file, made from R:
https://catalyst.uw.edu/sharespaces/download/17360/318803?html=1&url=https://catalyst.uw.edu/sharespaces/download/17360/318803
12 Aug 2012
Noticed this evening that some of the plates from the reinfection experiment look like they have developed lesions – especially for some of the blade inocula, it looks like there is a dark lesion that extends to either side of the inoculum. This morning, that was not as evident (at least, at a glance).
11 Aug 2012
Vibrio lab:
Dropped our larvae with 10% bleach and did final total counts:
| 1 |
2 |
3 |
4 |
Control |
|
| A |
21 |
42 |
51 |
44 |
35 |
| B |
32 |
40 |
34 |
45 |
32 |
| C |
34 |
34 |
33 |
55 |
41 |
Labyrinthula lab:
Made up new liquid cultures of several of the Laby cultures for the herbivore group:
- Sandy monoculture (1.17.1) à 500 µL of liquid culture into 10 mL of Laby broth
- BH Laby (Plate: “Beach Haven CK, 1. 7-18-2012, Replate 7-25-2012”) à 250 µL of liquid culture in 10 mL of Laby broth
- Scraping of plate “7/30/12 FB Sh-T-1 DH” in 10 mL of Laby broth
- Scraping of plate “FB SH T-1 7/30/2012” in 10 mL of Laby broth
- Scraping of plate “Shallow Bay 1 7.18.2012 CK” in 10 mL of Laby broth
Also replated several agar cultures that look like they’re starting to take over the plate. The False Bay samples were additionally replated (into separate plates) for the Herbivore group.
- Plate “FB SH T-1 7/30/2012”
- Plate “FB Sh T-1-D 7/30/2012 DH”
- Plate “Shallow Bay 1 7/18/2012 CK”
10 Aug 2012
Vibrio lab:
Plating results:
T1N5 plates from 8 Aug, with the dilutions of Vibrio, finally grew. Final counts:
| Dilution |
Rep 1 |
Rep 2 |
| 10^(-5) |
33 |
4 |
| 10^(-6) |
5 |
0 |
| 10^(-7) |
2 |
1 |
TBS plates also grew, but:
- It looks like there are contaminating bacteria on them.
- The bacterial density is pretty high—well over a couple thousand colonies per plate.
- The dilutions look as though they were done out of order: 10^(-6) > 10^(-4) > 10^(-5)
Larval bacterial challenge:
Because Ashton and I both had a lot else going on, we decided that we would take our 48-hour counts today, then drop the larvae and do the total counts tomorrow. We went over alive vs. dead with Carolyn—basically, what had been skipped was moving to a higher magnification when it looked like a larva was dead, to verify. I think it is likely that my assignment of what was dead was way too conservative; sometimes ciliates or bacteria make it look like the velum is vibrating at low magnification, but at high magnification it is clear that something is wrong. The numbers this time matched much more closely.
Once again, I counted row A and row C, and Ashton did the controls and row B.
Table of larvae mortality after 48 hours:
| 1 |
2 |
3 |
4 |
Control |
|
| A |
3 |
40 +/- 1 |
43 |
38 |
2 +/- 1 |
| B |
5 |
37 |
31 |
40 +/- 1 |
0 |
| C |
4 |
21 |
32 |
27 |
0 |
Notes:
- Well A1: “Broken shell now dead”
- Well A2: “One was moving, although I think it was because of the ciliate. Although the larva could have been eating it…”
- Well C2: “+3 not quite dead +2 that are turning in circles but otherwise look dead”
- Well A4: “May have counted one twice.”
Labyrinthula lab:
Reinoculation experiment:
We have two inoculation experiments going:
- Autoclaved blade inoculum (Protocol 1 under 9 Aug)
- Liquid culture inoculum (Protocol 3 under 9 Aug)
What we actually did in this experiment follows. A thorough protocol taking some of the problems into account will be posted later.
General:
- 2 L of seawater were sterilized by autoclaving. Because we wanted to use it the same day, we put it in an ice-water bath to cool it quickly.
- 32 empty petri dishes were labeled with an identifying number and a name, and they were filled with 25 mL of sterile seawater.
- Maya assigned the identifying number with the purpose of randomizing the locations of all plates on the table, rather than having plates from a particular treatment together.
- Blades that had been left in the seawater tank to heal were removed to a bag of seawater (for the interim). Each was:
- photographed with a ruler and an identifying number;
- dipped briefly in ethanol, then washed in sterile seawater;
- put into the corresponding plate with seawater until inoculation.
- NOTE: Even just brief dips in ethanol caused the blades to darken. This is obviously not ideal if we want to find lesions! Will try to locate another disinfecting agent that does not have this effect.
- After all inoculations had been performed, the plates were put into their randomized locations on the table by arranging them in numerical order (4 rows of 8). The plates that were not used were marked with an X and kept in the randomized formation.
- Thermometers were placed above and on top of the table to keep track of general temperature. (As of 14 Aug, they’ve remained stably at around 25 C.)
1. Autoclaved blade inoculum
Pictures of the inoculum blades.
Both the Sandy and 2* cultures had blades that looked infected; the 1* culture is still growing so it was dropped from this experiment and left to grow more. Sadly, there was contamination in one of the controls (#2)!
Control #1
Control #2
Sandy #1
Sandy #2
1*-1
1*-2
2*-1
2*-2
Replicates:
4 leaves total per plate à take 2 leaves and halve them
X 2 plates
X 2 cultures
+ 3 controls
This was dropped down to 2 leaf-halves per plate, for a total of 4 replicates per culture, + 3 controls = 11 clip inocula plates total
Procedure:
- Drew made new clips from a different kind of tubing. The pieces were put into 10% bleach briefly to sterilize them, then were transferred to distilled water, and finally to sterile seawater before the inocula were set up.
- Around 7pm, we set up an assembly line to put the blade inocula onto the fresh blades:
- I removed the autoclaved, infected blades from the petri dishes and cut them in half.
- For a couple of the inocula (plates TO BE ADDED), I used halves from two different blades because one did not look fully infected.
- I also noticed that the darker green leaves were much easier to take out of the agar without tearing them than the lighter green leaves—another reason those should not be used for autoclaved samples in the future!
- Instruments were sterilized briefly before going back into the same plate, and thoroughly between different plates. I also changed gloves between inocula from different cultures.
- Non-contaminated control plate was used for a non-infected control inoculum.
- Gregor took each blade inoculum and clipped it to the healthy blade.
- He changed gloves between every inoculum to prevent ANY contamination between samples.
- Sammy removed the clips from the sterile seawater and helped Gregor open them with the (thick) forceps, which both of them found to do better at that task than the more finely pointed forceps from the dissecting kits.
- Sandy sterilized the thick tweezers between each inoculum.
- I removed the autoclaved, infected blades from the petri dishes and cut them in half.
2. Liquid culture inoculum
Replicates:
Dilution factors 10^0, 10^(-2), 10^(-4)
X 3 reps per factor
X 2 cultures
+ 3 controls that would receive sterile seawater
Because the liquid culture from Sandy’s monoculture did not have enough cells/mL to make inoculations worthwhile, it was dropped from the experiment, and instead we did a full dilution series by factors of 10 from 10^0 to 10^(-4) for the BeachHaven culture, = 3 x 5 + 3 controls = 18 plates total.
Procedure:
10am
- Liquid cultures (BeachHaven and Sandy’s monoculture) were all vortexed well and combined. Because the total volume of culture was > 50 mL, they were each divided between two 50mL Falcon tubes (each tube containing ~ 33 mL of culture).
- Centrifuge that fit 50mL Falcon tubes was finally located—a very old one in the basement of Fernald. Culture lids were parafilmed and spun for 20-25 min on speed “5” (“Low”).
- Supernatant removed. Pellets resuspended in 15 mL Laby broth. Both Falcon tubes were combined to a single one.
- 10 mL of combined culture were removed to 15-mL Falcon tubes to use in inoculating new liquid cultures.
- Cultures in 50-mL Falcon tubes that had been resuspended in Laby broth were spun down again (5/ low) for 20-25 min.
- Supernatant removed. Pellets resuspended in 10 mL (autoclaved) sterile seawater. Ideally, these would have been used right away, to prevent the cells from being too badly damaged.
- Courtney counted the number of cells in each culture using a hemocytometer. Sandy confirmed what was Laby cell under the microscope. This was the point at which we realized that there were not enough cells in the monoculture to merit using/ diluting it, and the plates that we had labeled for that culture were reassigned to extra dilution factors for the BeachHaven culture. 3 plates were left as complete blanks (no treatment whatsoever).
- Calculations for the number of cells in the BeachHaven culture:
Hemocytometer counts in 10 µL of a 1:2 dilution of liquid culture:
| Rep |
Count |
Calculation |
Total |
| 1 |
48 |
48 x 2 x 10 x 1000 |
9.6 x 10^5 |
| 2 |
33 |
33 x 2 x 10 x 1000 |
6.6 x 10^5 |
| 3 |
36 |
36 x 2 x 10 x 1000 |
7.2 x 10^5 |
| Mean |
7.5 x 10^5 cells/mL |
Dilution factors:
10^0 dilution = (7.8 x 10^4 cells in 0.1 mL) / 25 mL = 3120 cells/mL
10^(-1) dilution = 312 cells/mL
10^(-2) dilution = 31.2 cells/mL
10^(-3) dilution = 3.12 cells/mL
10^(-4) dilution = 0.312 cells/mL
- Dilutions were made 100 µL of culture in 900 µL of sterile seawater and vortexed.
- 100 µL of dilution were added to the appropriate plate. The 10^0 plates received 100 µL of the full culture; the control plates received 100 µL of sterile seawater.
11 Aug 2012
Vibrio lab:
Dropped our larvae and did final total counts:
| 1 |
2 |
3 |
4 |
Control |
|
| A |
21 |
42 |
51 |
44 |
35 |
| B |
32 |
40 |
34 |
45 |
32 |
| C |
34 |
34 |
33 |
55 |
41 |
Labyrinthula lab:
Made up new liquid cultures of several of the Laby cultures for the herbivore group:
- Sandy monoculture (1.17.1) à 500 µL of liquid culture into 10 mL of Laby broth
- BH Laby (Plate: “Beach Haven CK, 1. 7-18-2012, Replate 7-25-2012”) à 250 µL of liquid culture in 10 mL of Laby broth
- Scraping of plate “7/30/12 FB Sh-T-1 DH” in 10 mL of Laby broth
- Scraping of plate “FB SH T-1 7/30/2012” in 10 mL of Laby broth
- Scraping of plate “Shallow Bay 1 7.18.2012 CK” in 10 mL of Laby broth
Also replated several agar cultures that look like they’re starting to take over the plate. The False Bay samples were additionally replated (into separate plates) for the Herbivore group.
- Plate “FB SH T-1 7/30/2012”
- Plate “FB Sh T-1-D 7/30/2012 DH”
- Plate “Shallow Bay 1 7/18/2012 CK”
9 Aug 2012
Vibrio lab:
Gram staining to identify whether an isolated population of bacteria is Gram-positive or Gram-negative.
- Liquid culture: Spread an inoculation-loop’s worth of culture on a glass slide.
- Allow water to air-dry.
- Pass the slide over a flame a few times to heat fix the bacteria to the slide.
- Place the slide on a rack over the sink (or other container) and flood the slide with crystal violet. Let stand for 1 min.
- Wash in a gentle stream of tap water to remove excess dye. Pass slide back and forth through the stream—you don’t want to wash off your bacteria!
- Flood with Lugol’s or Gram’s iodine for 1 min.
- Wash gently in tap water.
- De-colorize with alcohol-acetone solution: Squirt on some decolorizing agent, rinse immediately, squirt on more decolorizing agent, rinse immediately.
- Counterstain with safranin for 15 sec.
- Let air dry or blot very gently.
- Add a drop of immersion oil (for a 100x objective) and view under the microscope.
- We had a lot of roughly rod-shaped bacteria that were stained pink, so they were Gram negative bacteria.
Serum agglutination test
- On two depression slides (or two depressions of one slide), add:
- Control: 50 µL sterile seawater and 25 µL bacterial culture
- Test: 25 µL sterile seawater, 25 µL bacterial culture, 25 µL polyclonal antibody.
- Mix with a sterile pipet tip. Incubate for 5 min at room temperature.
- Compare the degree of agglutination under the microscope at 10x or 4x.
…I went through the protocol, but I got interrupted for other things before I could view the slides. By the time I got back to them, the liquid had evaporated, leaving me with a lot of salt crystals and some swimming bacteria in the control slide and a long, branching, somewhat snowflaked structure in the test slide. Try again tomorrow!
Oyster larval mortality, day 1
Counted the number of dead oyster larvae in the 12-well plates set up yesterday (8 Aug). Larvae were pronounced “alive” if a) they were clearly moving, or b) their cilia or velum were moving rapidly even if the larvae were stationary. Larvae in which there was no sign of motion, or in which there were sporadic “pulses” of motion, were pronounced “dead”. I was (perhaps) extremely conservative with my death counts: if any part of the cilia or velum looked like they were vibrating, I counted the larva as alive. Only when the velum was a) still or b) pulsing sporadically did I count the larva as dead.
Ashton counted the control wells and row B. I counted rows A and C.
Table of larval mortality after 24 hours:
| 1 |
2 |
3 |
4 |
Control |
|
| A |
3 |
6 +/- 1 |
7 +/- 1 |
12 +/- 1 |
1 |
| B |
2 |
20 |
21 +/- 1 |
28 |
2 |
| C |
1 |
6 +/- 1 |
4 +/- 1 |
7 |
0 |
Notes:
- Well A1: “Broken shell but still moving”
- Well A2: “Much less movement generally”
- Well B2: “Voided themselves perhaps?”
- Well C4: “Lots more movement and activity. At least one was probably dead at the start of the experiment.”
- I noticed that Ashton’s death counts were far higher than my counts for columns 2-4, so I looked at those wells too (to find out if it was a true difference or a counting difference). I counted 14 deaths in well B2; 9 in well B3; and 7 in well B4. So we seem to have some discrepancy in how we assign dead vs. alive. Hopefully this will get cleared up tomorrow with Caroline.
Plating results from the serial dilutions yesterday (8 Aug):
Sadly, nothing grew in the T1N5 or TBS plates. The T1N5 plates were parafilmed and moved to a 30 C incubator in hopes that they will grow a little faster. The TBS plates were left on the benchtop for another night.
The TCBS plate showed colony growth on the Vibrio side, but not on the Aeromonas side, demonstrating that the media is indeed selective for Vibrio growth. Colonies are fairly light colored.
Labyrinthulid lab:
Sandy’s Laby have grown well and are growing over 1-2 eelgrass. Our 2* cultures also appear to be taking off. Sadly, there is still nothing in the 1* cultures.
Pictures:
Sandy #1
Sandy #2
1*-1
1*-2
2*-1
2*-2
We discussed protocols for the reinoculations. We have 3 different approaches we could use:
- Inoculum from autoclaved blades (currently growing)—clip to healthy blades
- Dosed response: Inoculate autoclaved blades from (quantified) liquid culture, then clip to healthy blades.
- Dosed response II: Inoculate healthy blades directly with (quantified) liquid culture.
We decided to go for protocols 1 and 3 tomorrow.
Protocol 1:
- Gregor helped me cut healthy-looking blades from Sandy’s tank into approximately 8-cm pieces, and we accumulated around 35 of them. These have been left in a tank outside with flowing seawater running through it to allow the cuts to heal.
- Gregor also cut up some tubing that we will use as clips for the autoclaved-leaf inocula. These will be sterilized tomorrow before use by brief alternating dips in bleach, distilled water, and sterile seawater.
- Tomorrow, we’ll see how many inoculum leaves have been infected. We may cut leaves in half for more replication power.
- Photographs of all healthy blades will be taken so that we can distinguish previous blemishes from uncertain Laby infections.
- Inocula will be clipped to the healthy blades with the tubing clips and placed in a petri dish with sterile seawater. Each blade/inoculum combination will receive its own petri dish. We can use autoclaved leaves from the control plates as negative controls.
Protocol 2:
- Pool together all liquid cultures of the same type.
- Spin culture down to a pellet and resuspend in a smaller amount of seawater.
- Quantify 10 µL or so on the hemocytometer. Use this reading to determine dilutions for the dose test.
- Add healthy blades to sterile seawater in a petri dish (1 blade per dish) and pipet the desired dilution of Laby culture into the seawater.
Also to do tomorrow:
- Any Laby replatings that need to be redone.
- Restart Laby liquid cultures from the spun-down and resuspended cultures. Try establishing new liquid cultures from the existing ones. Start with a small volume and inoculate slowly into larger ones as the cultures grow.
Finally, we discussed running the PCR test on some of the Laby plates/ liquid cultures. Drew would like to run one of the molds from our field samples as a negative control for the primers. Be sure to discuss this with her!
8 Aug 2012
Labyrinthulid lab:
Reinfections:
Checked on the eelgrass infection plates from yesterday (7 Aug). Both of Sandy’s cultures are growing well. One 2* culture may have some growth, but it’s not certain that that particular culture is a labyrinthulid.
Pictures of each plate:
Control #1
Control #2
Sandy #1
Sandy #2
1*-1
1*-2
2*-1
2*-2
See 6 Aug for full names of the plates.
Vibrio lab:
Results of streaking from yesterday, 7 Aug:
- Plate 1:
- Half 1: Good line of Am; no appearance of Vibrio. Although it was streaked, it’s possible that no Vibrio actually made it onto the agar.
- Half 2: Streak looks good and has single colonies.
- Plate 2: Our Vibrio strain (X00123) grew on the TCBS plate.
Serial dilutions of Vibrio and Aeromonas cultures:
- Vibrio culture: RE66
- 1 mL of culture / previous dilution added to 9 mL of seawater and mixed thoroughly.
- Vibrio dilutions: 10^(-1) to 10^(-7)
- Aeromonas dilutions: 10^(-1) to 10^(-6)
| Media |
Bacteria |
Dilution |
Replicates |
| T1N5 |
RE66 |
10^(-5) |
2 |
| T1N5 |
RE66 |
10^(-6) |
2 |
| T1N5 |
RE66 |
10^(-7) |
2 |
| TSB |
Am |
10^(-4) |
1 |
| TSB |
Am |
10^(-5) |
1 |
| TSB |
Am |
10^(-6) |
1 |
Tomorrow we’ll use the colony counts on these plates to estimate the concentration of the initial cultures.
We also made a streak of the initial Am and RE66 cultures onto TCBS to test growth/ metabolism.
Plates inverted and incubated at room temperature overnight.
Inoculation of oyster larvae:
2 plates made: Experimental and control.
1. Oyster larvae added to all wells of the experimental plate and to 3 wells of the control plate.
- Ideal number of oysters: 40 +/- 5. Actual numbers of oysters (+/- about 2) are recorded in the table below.
- Oyster larvae were diluted from a concentrated stock. Between 25 and 300 µL of larvae were added to a few wells at a time in increments of either 100 µL (less concentrated dilution) or 25 µL (more concentrated dilution). Larval counts were verified under the dissecting scope.
3. Bacterial treatments: 100 µL of the appropriate bacterium at the appropriate concentration added to the wells of the experimental plate. Setup is recorded in the table below. Control wells did not receive any bacteria.
Calculations for dilution factors IN PHYSICAL NOTEBOOK, TO BE UPLOADED.
Setup of experimental plate. Notes:
- Columns 1, 2, and 3 contain replicate cultures with Vt only at different concentrations. This will evaluate dose response.
- Column 4 evaluates probiotic effect of Am on two concentrations of Vt. C4 contains the probiotic control with only Am.
- Control wells (on a separate plate, not shown in the experimental plate setup) contain only larvae and seawater.
| 1 |
2 |
3 |
4 |
|
| A |
10^4 RE66 = 10^(-3) dilution |
10^5 RE66 = 10^(-2) dilution |
10^6 RE66 = 10^(-1) dilution |
10^4 Am = 10^(-2) dil + 10^5 RE66 = 10^(-2) dil |
| B |
10^4 RE66 = 10^(-3) dilution |
10^5 RE66 = 10^(-2) dilution |
10^6 RE66 = 10^(-1) dilution |
10^4 Am = 10^(-2) dil + 10^5 RE66 = 10^(-1) dil |
| C |
10^4 RE66 = 10^(-3) dilution |
10^5 RE66 = 10^(-2) dilution |
10^6 RE66 = 10^(-1) dilution |
10^4 Am = 10^(-2) dil |
Initial larval counts per well:
| 1 |
2 |
3 |
4 |
Control |
|
| A |
34 |
41 |
~56 |
~55 |
40 |
| B |
33 |
44 +/- 2 |
36 |
41 |
32 |
| C |
38 |
34 |
34 |
55 +/- 2 |
41 |
Note for the broader experiment: Each group has a set of plates to measure, in total the effects of
- Vt RE022 at ambient pCO2
- Vt RE022 at 2000 µM pCO2
- Vt RE98
- Vt RE101
- Vt RE66
Microplate cultures allowed to grow overnight at room temperature.
7 Aug 2012:
Labyrinthula lab:
Reinfections:
From each of the BeachHaven CK, DH replate, and Sandy’s Laby cultures (first three plates from 6 Aug), 2 small slabs of agar were cut from the leading edge with sterile instruments and transferred to 2 fresh plates. 4 pieces of autoclaved eelgrass were then arranged around the agar cut in a roughly rectangular fashion. 2 plates served as contamination controls, and received eelgrass but no agar cut. Pictures:
Note that although some of the plates are dated 8/6, all plates were in fact made today.
Also, a note for the future: I had cut up and autoclaved both young and old parts of the blades. It turns out that the younger (thinner, lighter green) segments do not do as well in the autoclave. Most of those pieces disintegrated, so they were used as controls. Next time, using exclusively older parts of the blade will prevent this problem.
Plates were parafilmed and put in Sandy’s lab with the heater running on low to provide a little extra heat for growth.
Vibrio lab:
Streaking practice from stock plates of Vibrio tubiashii (Vt) and Aeromonas media (Am).
Ashton’s and my strain: X00123
1. T1N2 plates:
Half 1: Probiotic test, a curved streak of Vt overlaid by a line of Am.
Half 2: Streak test: Vt streaked out to single colonies.
See diagram below.
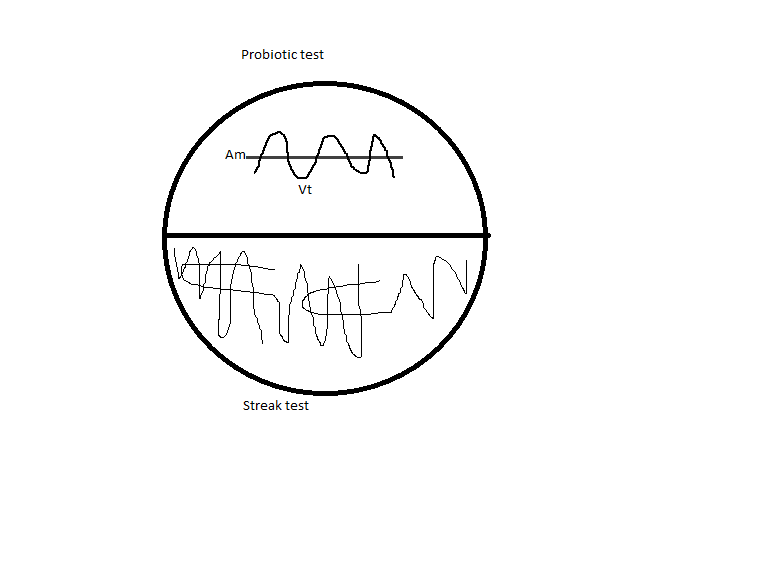
2.TCBS plates: Selective for Vt; depending on the metabolism of the colony, it will turn a different color.
One TCBS plate was divided into six portions, and each member of the class made a streak from our Vt stock plate.
Protocol for spec pH:
- Turn on the spec light source at least 1 hour before the first reading.
- Sample from the bottom of the water you want to pH measure. This avoids contact with atmospheric gases.
- Take 50 mL of water with a 50-mL serological pipet. Load 20 mL into the cuvette. Note: Avoid touching the round ends of the cuvette.
- Add the sample to a bag of water (with sample info) in the 25 C bath for at least 20 min.
- Make sure the Ocean Optics box is plugged into the computer.
- In SpectroSuite, make sure the settings are one 5 ms, 50 boxcar, and 3 strobelamp.
- Blank the spec in SpectroSuite:
- Click the “light” button.
- Flip the spec light switch and click the “light” button again.
- Ask for absorbance by clicking “A” in SpectroSuite.
- Enter the temperature of the sample to the Excel spreadsheet. Use the CO2Calc for any necessary back-calculations to the source water.
- Forward to a tab in the Excel spreadsheet that does not have calculations. In the sheet:
- Blue = background
- Purple = pH
- Maroon = slope for dye correction—we will not need this
- Take the measurement:
- Remove the cuvette from the bath and dry with LENS PAPER ONLY.
- Insert the cuvette into the holder with the letters facing forward.
- Replace the lid of the holder.
- Press “copy” in SpectroSuite and paste it into the blue column of the Excel spreadsheet.
- Remove the cuvette and uncap one of the lids.
- Add 70 µL of dye to the cuvette, inserting the pipet as far into the center of the cuvette as possible. Mix in the dye by inverting.
- Replace the cuvette in the holder, letters facing forward.
- Press “copy” in SpectroSuite and paste the values into the purple column of the Excel spreadsheet.
- Copy the results of the blue and purple columns (to the right, also color coded) from the Excel sheet and paste “Values only” to the first page of the spreadsheet.
- Measure the salinity of the sample with the salinometer.
6 Aug 2012
Vibrio lab:
Experiment preparation:
- Filled and autoclaved 15-mL tubes with 9 mL of DI water for serial dilutions
- Started cultures of Vibrio growing at ambient pCO2 and 2000 µM pCO2
Went over the protocol for taking alkalinity measurements:
- Run everything preferentially at 25 C (industrial standard), but above all at the same temperature.
- Allow the machine and water baths to warm up for 15-30 min.
- Uncap the pH probe. Ensure that there is electrode filling solution in the lower cap, and in the probe to the first line.
- Make sure the bottle of HCl is at least half full. Swirl briefly to homogenize.
- NOTE: Use gloves when handling the bottles with the black stoppers—they contain mercury!
- Calibrate the pH probe.
- Set the sensor (probe type, DG115).
- Press Modify à Calibrate à Modify (to check buffers) à OK.
- Rinse the probe with DI water and blot dry. Place the probe in the circular stand next to the rotor.
- Pour the (pink) 4.0-pH solution into a cup. Place the cup in the Styrofoam insulator and attach to the circular holder.
- Press OK to read the solution.
- Remove the cup and rinse the probe in DI water.
- Pour the (yellow) 7.0-pH solution into a cup. Place the cup in the Styrofoam insulator and attach to the circular holder. Press OK to read the solution.
- Remove the cup and rinse the probe in DI water.
- Pour the (blue) 10.0-pH solution into a cup. Place the cup in the Styrofoam insulator and attach to the circular holder. Press OK to read the solution.
- Remove the cup and rinse the probe in DI water.
- The resulting output of mV/pH should have a slope between (-)55 and (-)59. If not, switch out the 10.0-pH solution and try again.
- Remove the cup and rinse the probe in DI water.
- Hit Save.
- Purge the burette.
- Press Burette.
- Select the option to rinse for 3 cycles. Insert a cup to catch the waste acid.
- Press Start.
- Dump the waste in the bottle labeled “HgCl2”. Rinse the probe.
- Check standard solution.
- Hit Reset and choose Alkalinity Method 2 (F2).
- Weigh out approximately 50g of solution CRM into a dry cup. Make sure there are no drops on the side of the cup.
- Put the cup + insulator into the circular holder. Enter the weight to the screen and press OK.
- Compare the output alkalinity with the corrected alkalinity via the equation: (Vol titrant) * (Conc. titrant) * (mass CaCl2) * 10^5 / (mass sample)
- Rinse the probe with DI water.
- Repeat measurement 3-5 times to ensure machine accuracy, or to use in a correction: (Average value) / (True value) * (Sample value)
- Measure sample in the same way as the standard.
- Clean up:
- Throw gloves in the Hg waste bin.
- Rinse and blot probe, recap, and return to storage cap.
- Rinse used cups three times in DI water.
Labyrinthula lab:
Reinfections:
As a Stage 1, we will add autoclaved eelgrass to Labyrinthula plates and see if the Laby can infect the eelgrass. I cut putatively healthy blades of eelgrass from the tank cut into 3-cm pieces and autoclaved them. Tomorrow, slabs of agar with Laby will be put into new plates with 4 blades of eelgrass. We will have two plates each of:
- BeachHaven CK 1 replate, feathery (3 Aug) (1*)
- False Bay DH replate (5 Aug) (2*)
- Sandy’s culture (#1)
- Sandy’s culture (#2) / Control with no Laby
Image of the game plan I put on the board (courtesy Courtney):
5 Aug 2012
Labyrinthulid lab:
The replate of Drew's culture (4 Aug) and the replate of the "feathery" section of one of Catherine's cultures (3 Aug) are both showing signs of growth. The other plates (from 3 Aug) are not. The agar chunks in these plates seem to be very wet--perhaps a problem with moisture? There may be something growing on the water, but it's hard to tell, even under the microscope.
4 Aug 2012
Labyrinthulid lab:
Located the good culture that Drew had made from a False Bay plant; because it seemed to be growing quite a bit, made a replate from part of the leading edge.
The room-temperature cultures for inoculation that Lisa had made (3 Aug) were in the sun, at a tangible temperature difference from parts of the room that were not exposed to sun. I moved the cultures to a bench top instead.
3 Aug 2012
Labyrinthulid lab:
Gel electrophoresis of the PCR products from 2 Aug. Visualization on a 1.5% agarose gel by SybrSafe using UV illumination. Because we had problems with the last ladder, a different, more reliable ladder was used instead. We used 7 µL of each PCR product and 5 µL of ladder.
Order of wells:
| Comb 1 |
Comb 2 |
||
| Well number |
Sample |
Well number |
Sample |
| 1 |
Jamie’s sample |
1 |
Pos. control for wells 3-4 |
| 2 |
Maya’s sample |
2 |
Neg. control for wells 3-4 |
| 3 |
Sonia’s sample (2 µL DNA) |
3 |
Jenna’s sample |
| 4 |
Pos. control for wells 1-3, 6 |
4 |
Ana’s sample |
| 5 |
Neg. control for wells 1-3, 6 |
||
| 6 |
Sonia’s sample (5 µL DNA) |
||
| 7 |
Neg. control for wells 7-15 |
||
| 8 |
PCH-12 |
||
| 9 |
DC1-D12 |
||
| 10 |
PD1 |
||
| 11 |
PD2 |
||
| 12 |
LD1 |
||
| 13 |
LD2 |
||
| 14 |
LH |
||
| 15 |
PH |
||
| 16 |
Pos. control for wells 7-15 |
||

Results:
- Ladder looks better (individual bands are at least distinct), but it is still running oddly.
- All positive controls (wells 4 and 16 in the first row, well 1 in the second row) show a band of approximately 500 bp.
- All negative controls (wells 5 and 7 in the first row, well 2 in the second row) are blank (no bands).
- The bands for both of my samples (wells 3 and 6 in the first row) show a band of the same length as the positive control, which is reassuring!
- DNA isolated from eelgrass (wells 3, 4 in second row; wells 1, 2, 8, 9 in first row) all show bands at about 500 bp. This is regardless of the (putative) health state of the eelgrass, indicating that Laby is ubiquitous.
- Well 9 (first comb) appears to have a double band.
- DNA isolated from snails (wells 10, 11) and Phyloplesia (wells 12-15) that had been feeding on diseased/ healthy eelgrass show bands, but perhaps at different lengths—especially for DNA isolated from snails.
- Wells 10, 11, and 14 appear to have a double band. 12 and 13 may have faint second bands.
- Well 15, a healthy Phyloplesia, has a band of distinctly different length from the other (disease-fed) Phyloplesias.
- Suggests that Laby may also be ubiquitous in herbivores. However, note that these animals had been in a tank with diseased eelgrass, and had possibly been feeding on it before the experiment. They were only fed on the explicitly diseased/ healthy eelgrass for 18 hrs before being dissected.
A BLAST search with the primer sequences showed some hits for other (unicellular) organisms beside Laby, but nothing in particular that popped out. In addition, all hits for non-Laby species were only with the forward primer. The combination of forward AND reverse primer sequences ONLY appears in Laby species.
Ideally, if we wanted to find out why these bands are different, we would want to either extract the bands or re-do the PCR and send the product in for sequencing.
Inoculation experiment
Discussed protocols for the inoculation experiment further. Separate cultures of Laby are currently being grown at room temperature and 37 C.
Literature protocol: From Muehlstein LK, Porter D, Short FT. 1988. Labyrinthula sp., a marine slime mold producing the symptoms of wasting disease in eelgrass, Zostera marina. Marine Biology 99:465-472.
Day 1
- Place plants to be inoculated into a 2-liter Erlenmeyer flask (or hollow plastic tube) and allow to acclimate for 48 hours. Literature conditions:
- Aerated artificial seawater at 36% salinity OR 0.45µM Millipore-filtered natural seawater at 36% salinity.
- 300 µE/(m*s) of illumination on a 14h light : 10 h dark cycle
- Constant temperature of 17 C
- Autoclave small (1-cm) pieces of healthy eelgrass in seawater. Place the pieces on an agar surface containing Labyrinthula. (In the paper, yeast acted as a food source for the Labyrinthula cultures. The authors report that the Labyrinthula would grow away from the yeast after 2-3 days.)
- Allow live plants to continue to acclimate.
- Allow Labyrinthula to invade the sterilized pieces of eelgrass.
- Once the sterilized eelgrass leaves have been invaded, perform the inoculations within 2-4 days:
- Clamp a single invaded leaf piece to a healthy, living eelgrass shoot 1-2 cm above the sheath.
- Monitor every 3-4 hours for the first 24 hours, then daily for the appearance of disease symptoms.
- Control plant: Clamp an autoclaved leaf from a clean petri dish (no Labyrinthula).
- Use imaging software to compare lesion sizes in inoculated and control plants.
- Optional: Re-isolate Labyrinthula from the lesions by plating on SSA.
Lisa’s protocol:
Day 1-2
- Grow up Laby cultures at desired temperatures
- Cut a putatively healthy blade of eelgrass into pieces of 2cm (possibly more). Leave in the seatable to allow cuts to heal.
- Vortex the Laby cultures.
- Use a hemocytometer to gain some indication of culture densities.
- Based on the density of the full culture, make serial dilutions of 10x or 100x. OR pellet the Laby cells and resuspend to desired density in seawater.
- Sterilize the eelgrass pieces and place them on a petri dish (with some seawater?). Perhaps scratch or provide another form of mechanical stress as a point of entry for the Laby.
- Squirt a set volume of resuspended/ diluted Laby culture onto each blade piece. Control set would receive a squirt of seawater with no Laby.
- Allow to grow/ infect.
- Record presence/ absence of lesions. Use imaging software to analyze the size of lesions in each treatment.
We discussed doing this with old blades vs. young blades (is there a difference in susceptibility?). Sammy suggested mixing the Laby in with the agar and allowing it to infect the blades in that way. There was some concern that an experiment like this would not have real ecological relevance, especially as regards the variables we measured in the field, but we seem to have decided to pose it as:
1) a test of Koch’s postulates (can we get reinfection),
2) a new method for inoculation, and
3) a way to test some basic hypotheses.
We would like to use both verified Laby samples from Sandy and a Laby sample that we isolated from the field.
Laby culture transfers
Lisa made new cultures in 50-mL Falcon tubes as well as in Petri dishes (to provide greater surface area, if that is better for Laby growth) and put them at 25 C and 37 C. I transferred, to separate agar plates, a block of agar from some restreaked plates:
- “Parent” plate from which I took the re-streak sample 31 Jul
- “Feathery” Laby from the re-streaked plate (made 31 Jul)
- “Smooth” Laby from the re-streaked plate (made 31 Jul)
2 Aug 2012
Morning: Resampling of eelgrass at False Bay.
- Added a third shallow transect.
- Re-did all quadrats in the shallow transects.
- Completed transects and quadrats for 3 deep transects.
- All other protocols as per 1 Aug.
As compared to Picnic Cove and Beachcomber Bay, False Bay seems to have generally less dense / more patchy eelgrass in the areas we surveyed. Shallow transect seemed to show smaller individuals and less disease than deep transects.
Afternoon: PCR on the DNA extracted 1 Aug.
Reactions:
- Jamie’s sample
- Maya’s sample
- My sample (2 µL DNA)
- Positive control (provided by Lisa)
- Negative control (water)
- My sample (5 µL DNA)
Stock mix:
| Reagent |
µL/ reaction |
X 6 = |
µL/ stock |
| Go-Taq 2x master mix |
12.5 |
75 |
|
| Primer 1 |
0.8 |
4.8 |
|
| Primer 2 |
0.8 |
4.8 |
|
| BSA |
1.5 |
9 |
|
| Nuclease-free H2O |
7.4 |
44.4 |
|
| Total |
23 |
138 |
Mixture was aliquoted in 23-µL increments into PCR tubes and 2 µL of the appropriate DNA (/water) were added to each tube.
I made up a separate PCR tube for my reaction with 5 µL of template:
| Reagent |
µL/ reaction |
| Go-Taq 2x master mix |
12.5 |
| Primer 1 |
0.8 |
| Primer 2 |
0.8 |
| BSA |
1.5 |
| Nuclease-free H2O |
4.4 |
| DNA |
5 |
| Total |
25 |
Primers:
Forward: Laby WC F
3’ GGAAGGCAGCAGGCGCGTAA 5’
Reverse: Laby WC R
3’ GAATGTTTCCAATCCTCAGTCGGTATAG 5’
Cycling conditions:
| Step |
Temperature ( C) |
Time |
| 1 |
95 |
10 min |
| 40 cycles of: |
||
| 2 |
95 |
30 min |
| 3 |
50 |
30 sec |
| 4 |
72 |
60 sec |
| 5 |
72 |
10 min |
PCR products stored overnight at 4 C.
1 Aug 2012
Labyrinthulid lab:
Morning: Sampling of eelgrass in Picnic Cove. See protocol for 30 Jul.
- 3 low-tide (“deep”) and 3 high-tide (“shallow”) transects measured.
- Eelgrass 6 inches to either side of the transect included.
- Density readings included egg masses and herbivores.
Picnic Cove had extremely muddy water, and most eelgrass measured (in both the shallow and the deep transects) had lesions on them.
Afternoon:
Transmission experiments
Discussed with Sandy some alternatives to the mesocosms for running transfection experiments. We will most likely be using small, upright cylinders in the quarantine room to prevent infections from entering the bay with outflow water. Ashton suggested several ways to attach the cylinders easily to a base. We can also put small heaters into some of the cylinders to test the effect of temperature, and tidbits to record temperatures.
The transmissions will most likely be done as per [name of paper]: autoclave a section of eelgrass, infect in with Laby on a plate, and then clip in to a healthy, living eelgrass and count the number of lesions that develop after 4-5 days.
PCR on Laby
Used eelgrass samples previously collected as substrates for DNA extraction and amplification. See 26 Jul for protocol.
I extracted DNA for a positive control from Laby grown on plates:
- Plate: Beach Haven CK
- Plate examined under dissecting scope to ensure that the organism growing looked like Laby.
- Streak taken from the leading edge (location marked on plate underside) and mixed into 700 µL of Buffer ASL.
- The rest of the protocol followed as per an extraction from tissue.
- The 1-minute incubation period after the addition of InhibitEx was accidentally skipped; however, since there was not much tissue to begin with (it would only be tissue from the Laby cells), this should not have much effect on the final product.
- Because the final elution was in 100 µL of Buffer AE and there was not much material to start with, it is possible that the elution does not have a very high concentration of DNA. Perhaps try two PCR reactions with 2 µL and 5-10 µL?
Data entry
Transcribed the data sheets that Amy and I recorded this morning into the giant spreadsheet:
https://docs.google.com/folder/d/0B5AIgISTKSN1V19DMDBObG8yN3c/edit?pli=1&docId=0ApAIgISTKSN1dHVJeUI1MWNSWjc5MlV4clZsRV9oUHc
31 July 2012
Labyrinthulid lab:
Morning: Sampling of disease prevalence in Beachcomber/ Beach Haven on Orcus. Modifications from the sampling protocol from 30 Jul:
- 2 low-tide (“deep”) transects and 2 high-tide (“shallow”) transects measured.
- For the deep transects, measurements were taken for 2.5m stretches, since the total blade density was much higher than at False Bay.
- Only eelgrass right along the transect were measured rather than to 6 in. on either side (for all transects).
- Flowering eelgrass were not counted.
- Herbivore counts were added to the density counts in the transects.
Afternoon:
Game plan for what we can do with the rest of the lab:
- Field sampling: False Bay, Beachcomber, Picnic Cove à already well underway.
- Inoculation experiments: in a mesocosm or smaller environment. Possibility of crossing the inoculations with a variety of temperatures and/or herbivore presence.
- PCR on laby. Try isolating from herbivores as well.
Culturing Laby
Plated lesion sections from 6 plants collected today. Plates labeled with “7/31, BC” and the plant identification number. Key: SH1 = shallow transect 1; SH2 = shallow transect 2; D2 = deep transect 2; -# = plant ID number. Note that SH1 only has plants #1 and 4.
Photos:
Healthy sections of the same plants were put into desiccating agent for Sandy’s collection for microsatellite analysis.
I also replated (streaked) an older plating on which a fungus was growing (Plate: “Beach Haven CK, 1. 7-18-2012, Replate 7-25-2012”) onto a new plate (labeled “7/31 SS, Replate of Beach Haven CK”). Because there were two non-fungal growths, I took a streak of each one on separate halves of the plate and labeled them on the plate bottom (as “smooth” or “feathery”).
PCR on herbivores
Helped Sammy set up cages with an herbivore on infected or uninfected eelgrass blades from Sandy’s tank.
30 July 2012
Labyrinthulid lab:
Morning: Sampling of disease prevalence of Laby at False Bay. Link to complete sampling methods TO BE INSERTED. In brief:
- Because the tide was coming in, three high-tide (“shallow”) transects were taken and no deep transects.
- Transects were measured for 10m total in two 5m stretches.
- Eelgrass shoots along the transect and for 6 in. on either side were measured for:
- Total length
- Presence of Laby lesions
- Length of each Laby lesion
- Lesions were identified conservatively—brown, necrotic area surrounded by a black edge. Any “grey area” identifications were not counted.
- Quadrats along each transect were used to compile density measurements of eelgrass.
Afternoon: Histology of Laby
Histology slides from diseased and non-diseased eelgrass from Picnic Cove had already been made up by Catherine. We were to provide independent analyses of the diseased state of the eelgrass by quantifying the number of Laby cells in each one.
Method:
- Evaluate 3 “diseased” slides and 1 “healthy” slide.
- Count the number of Laby cells per view in 5 views at 40x magnification.
- Identification of Laby cells was conservative—unless it had BOTH the spindle shape AND evidence of some sort of mucus, it was not counted.
- For slides with more than one blade sample on them, individual blades were evaluated and numbered from left to right (when the slide label is facing left).
Counts for the slides I looked at can be found in the Google Docs spread.
Culturing Laby
Purpose: Culture Laby for PCR, re-inoculation, etc.
Method:
- Sterilize instruments by flaming them with ethanol.
- Choose a piece of eelgrass with a good lesion and photograph with its identification name and a ruler for scale/ size of lesion.
- Cut out a section from the lesion that is otherwise fairly clean.
- Dip section in ethanol briefly and let dry.
- Cut section into 3-4 smaller pieces and arrange on a plate with Laby media.
- Let incubate for two days at room temperature.
Photo of the culture:
27 July 2012
Armina Lab
Visualization of the PCR products made yesterday (26 Jul) on a 1.5% agarose gel, visualized with SybrSafe and UV illumination.
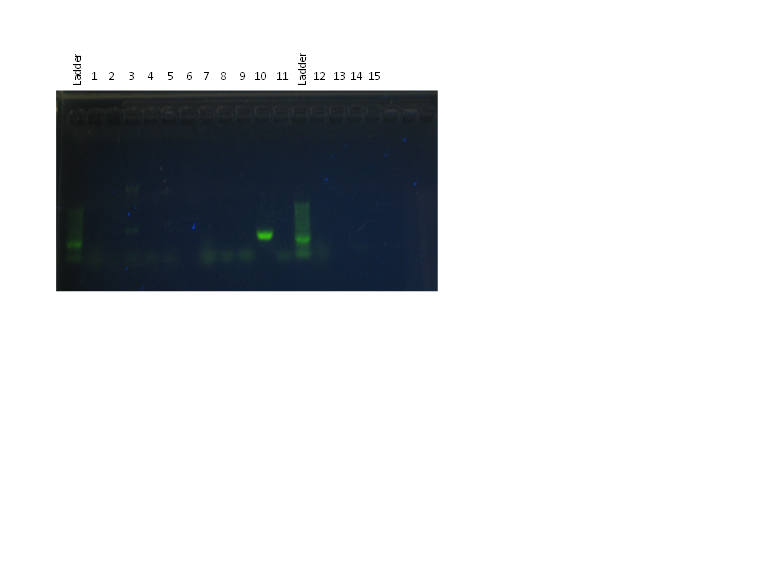
Order of wells (see 26 Jul for details of samples and primer sets):
| Number |
Primer set |
Sample |
| 1 |
Universal bacterial |
My sample |
| 2 |
Ashton’s sample (well punctured) |
|
| 3 |
Positive control |
|
| 4 |
Negative control |
|
| 5 |
Ashton’s sample (re-load) |
|
| 6 |
Empty |
|
| 7 |
Ehrlichia |
My sample |
| 8 |
Ashton’s sample (well punctured) |
|
| 9 |
Ashton’s sample (re-load) |
|
| 10 |
Positive control |
|
| 11 |
Negative control |
|
| 12 |
WS |
My sample |
| 13 |
Ashton’s sample |
|
| 14 |
Positive control |
|
| 15 |
Negative control |
Results:
- The ladder did not work well and was smeary—this was a problem for the entire class.
- All positive controls (wells 3, 10, 14) showed bands. 3 has two bands, both over 1500 bp; 10 has a bright band at ~2000 bp; and 14 has a faint band at ~1500 bp.
- Negative controls only show evidence of primer dimers, no contamination.
- All other wells have only primer dimers. In the case of my sample, this may be because the specimen was not actually infected. It is also possible that the particular primer set used did not anneal to any DNA from the putative infection.
- Lisa mentioned afterwards that it might have been a good idea to run a primer set for eukaryotic DNA, since that most likely forms the vast majority of DNA in our tissue samples.
26 July 2012
Armina Lab: DNA extraction and PCR
DNA extraction: QIAGen Stool kit
1. From the tissue samples stored in ethanol yesterday (25 Jul; acc 12-1-5), one piece of tissue was extracted using sterile dissecting tools, minced into small pieces, and placed into a 2-mL microcentrifuge tube. (The sample should be weighed before being minced, but the lab scale was not sensitive enough to record such a small mass.)
Break up tissue cells.
2. 700 µL of Buffer ASL added to the microcentrifuge tube and the contents vortexed for 1 min. Then a second set of 700 µL Buffer ASL was added and the contents vortexed until the mixture was roughly homogenous. (Note: Tube vortexed for several minutes without complete homogenization, but the quantity of tissue in solution did increase over that time.
3. Mixture heated for 5 min at 70-75 C
4. Sample vortexed briefly, then centrifuged at full speed for 1 min.
5. 1.2 mL of the supernatant was pipetted into a new 2-mL microcentrifuge tube.
Remove PCR inhibitors.
6. 1 InhibitEX tablet added to the sample and vortexed until the tablet had dissolved. Tube incubated for 1 min at room temperature.
7. Sample centrifuged at full speed for 3 min. Supernatant transferred to a new microcentrifuge tube and re-centrifuged for 3 min at full speed.
Digest cellular proteins.
8. A new microcentrifuge tube was set up with 15 µL of Proteinase K. 200 µL of the resultant supernatant from Step 7 was transferred into this tube, and 200 µL of Buffer AL was added.
9. Sample + Proteinase K + Buffer AL vortexed for 15 sec.
10. Tube incubated for 10 min at 70 C.
Wash steps.
11. Samples centrifuged briefly.
12. 200 µL of 95% molecular-grade ethanol added to the tube, vortexed, and spun down.
13. Mixture applied to a QIAamp spin column and centrifuged for 1 min at 8000 rpm.
14. Column transferred to a new collection tube and 500 µL of Buffer AW1 were applied. Column spun for 1 min at 8000 rpm.
15. Column transferred to a new collection tube and 500 µL of Buffer AW2 were applied. Column spun for 3 min at full speed (~16.3 rpm).
Elution.
16. Column transferred to a clean 1.5-mL microcentrifuge tube and 100 µL of Buffer AE were applied. Column incubated at room temperature for 1 min, then spun for 1 min at 8000 rpm.
17. Column discarded; elution stored at 4 C.
PCR of DNA samples to determine presence/ type of DNA from a disease-causing agent.
We are running three PCR reactions on our armina DNA samples:
| Name |
Primer set |
Protocol |
Detection |
| Universal |
EUB A / EUB B |
Generic |
Any bacteria |
| Ehrlichia |
EHR16s F / EHR16s R |
Generic |
Rickettsia |
| WS |
RA 3-6 / RA 5-1 |
WS-RLO |
Particular Rickettsia species from abalone |
Reactions: (for each reaction set)
- My sample (12-1-5 DNA extracted 25 Jul)
- Ashton’s sample (a stock sample from a previous year)
- Positive control (provided by Lisa)
- Negative control (water)
Generic protocol:
Stock solution:
| Reagent |
µL/ reaction |
X 5 = |
µL/ stock |
| Go-Taq 2x master mix |
12.5 |
62.5 |
|
| Primer 1 |
0.8 |
4.0 |
|
| Primer 2 |
0.8 |
4.0 |
|
| BSA |
1.5 |
7.5 |
|
| Nuclease-free H2O |
7.4 |
37.0 |
|
| Total |
23 |
115 |
Two sets of Generic stock were made up, one with the EUB A/ EUB B primer pair, the other with the EHR16s F/ EHR16s R primer pair.
23 µL from each stock were pipetted into 4 PCR strip tubes, and 2 µL of the appropriate DNA sample (marked above as reactions) were added to each tube.
Cycling conditions:
| Step |
Temperature ( C) |
Time |
| 1 |
95 |
10 min |
| 45 cycles of: |
||
| 2 |
95 |
15 sec |
| 3 |
60 |
1 min |
WS-RLO protocol:
Stock solution:
| Reagent |
µL/ reaction |
X 5 = |
µL/ stock |
| Go-Taq buffer |
4.0 |
20.0 |
|
| Primer RA 3-6 |
0.1 |
0.5 |
|
| Primer RA 1-5 |
0.1 |
0.5 |
|
| BSA |
0.8 |
4.0 |
|
| dNTPs |
0.4 |
2.0 |
|
| MgCl2 |
1.2 |
6.0 |
|
| Taq polymerase |
0.32 |
1.6 |
|
| Nuclease-free H2O |
11.08 |
55.4 |
|
| Total |
18 |
90 |
18 µL of stock were added to each of 4 PCR strip tubes, along with 2 µL of the appropriate DNA sample (reactions above).
Cycling conditions:
| Step |
Temperature ( C) |
Time |
| 1 |
95 |
3 min |
| 40 cycles of: |
||
| 2 |
95 |
1 min |
| 3 |
62 |
30 sec |
| 4 |
72 |
30 sec |
| 5 |
72 |
10 min |
Gels of all sets will be visualized tomorrow.
25 July 2012
Armina lab: Dissection of Armina californicum:
Accession number: 12-1-5
Total mass: 4.3 g
Total length: 36.4 mm
General observations:
- Wood-brown back with creamy, rather broken stripes
- Foot is a reddish-brown color
- Wing edges are also a creamy color
- Two potentially abnormal areas noted in the diagrams below:
(Note on the previous dissection: Animals had been used for neuroethology, so they had already undergone one operation to take measurements and samples from the brain.)
Digestive gland was extracted from a small transversal cut on the dorsal side behind the head and removed from the connective tissue. The digestive gland was cut transversally in three places to create four pieces, as pictured below:

Each of the pieces was cut in half:
- One half was put into a quadrant of the histological cassette;
- The second half was put into a PCR tube containing 1 mL of 95% molecular-grade ethanol.
The histological cassette was immersed in Invertebrate Davidson’s fixative for 24 hours. The PCR tube was stored at 4 C.
24 July 2012
Invertebrate dissections:
Oyster:
Shell cracked open at hinge and pried apart; adductor muscle cut from the right-hand shell to expose the oyster in the left-hand shell. Two cross-sections were taken width-wise across the oyster, just above the labial palps. The heart was also exposed.
Diagrams from the dissection:
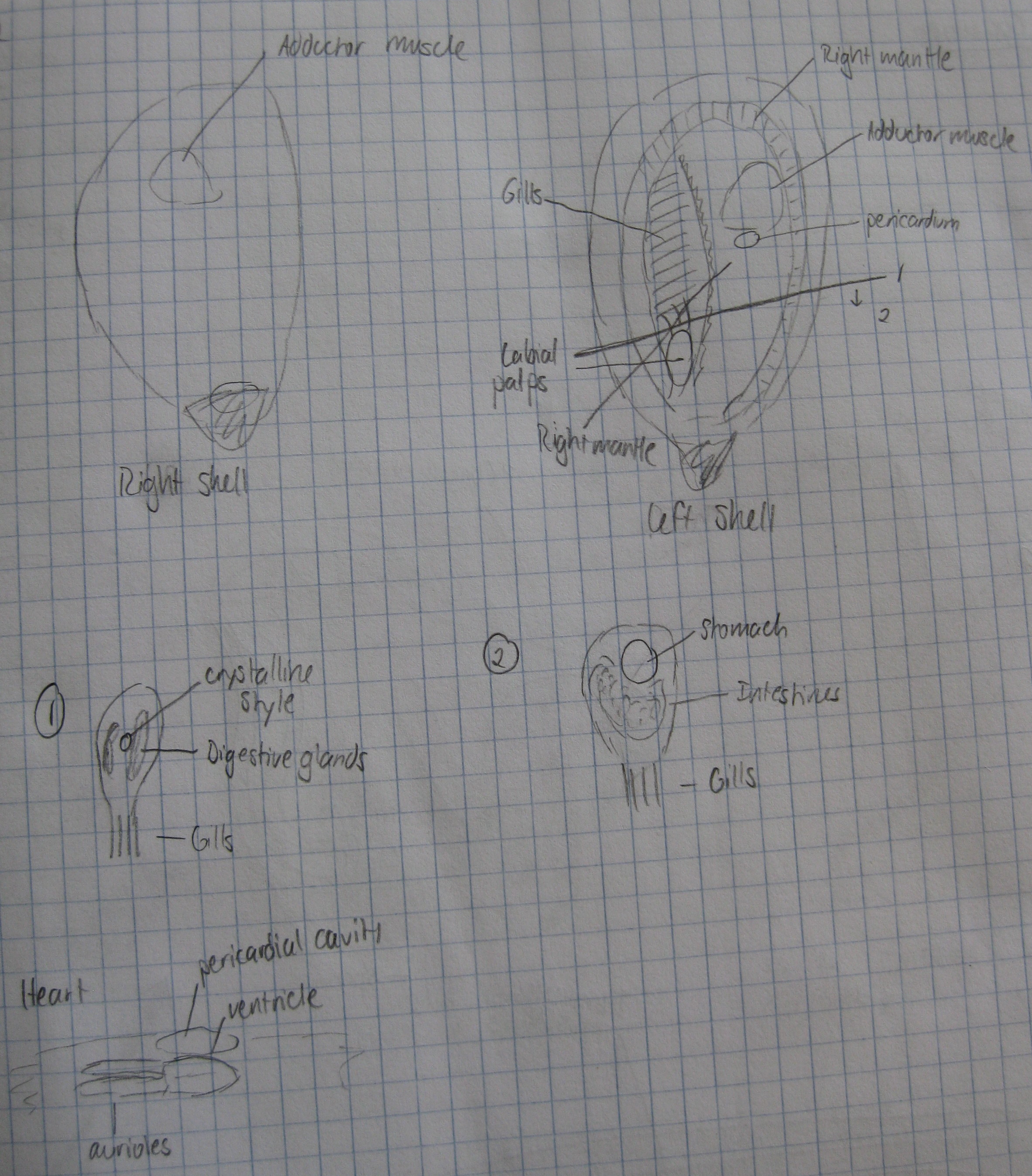
Sea cucumber:
Organism was cut lengthwise along the tentacles, starting with the aboral end. Most of the internal organs spilled out before the cut was very large.
Diagrams:
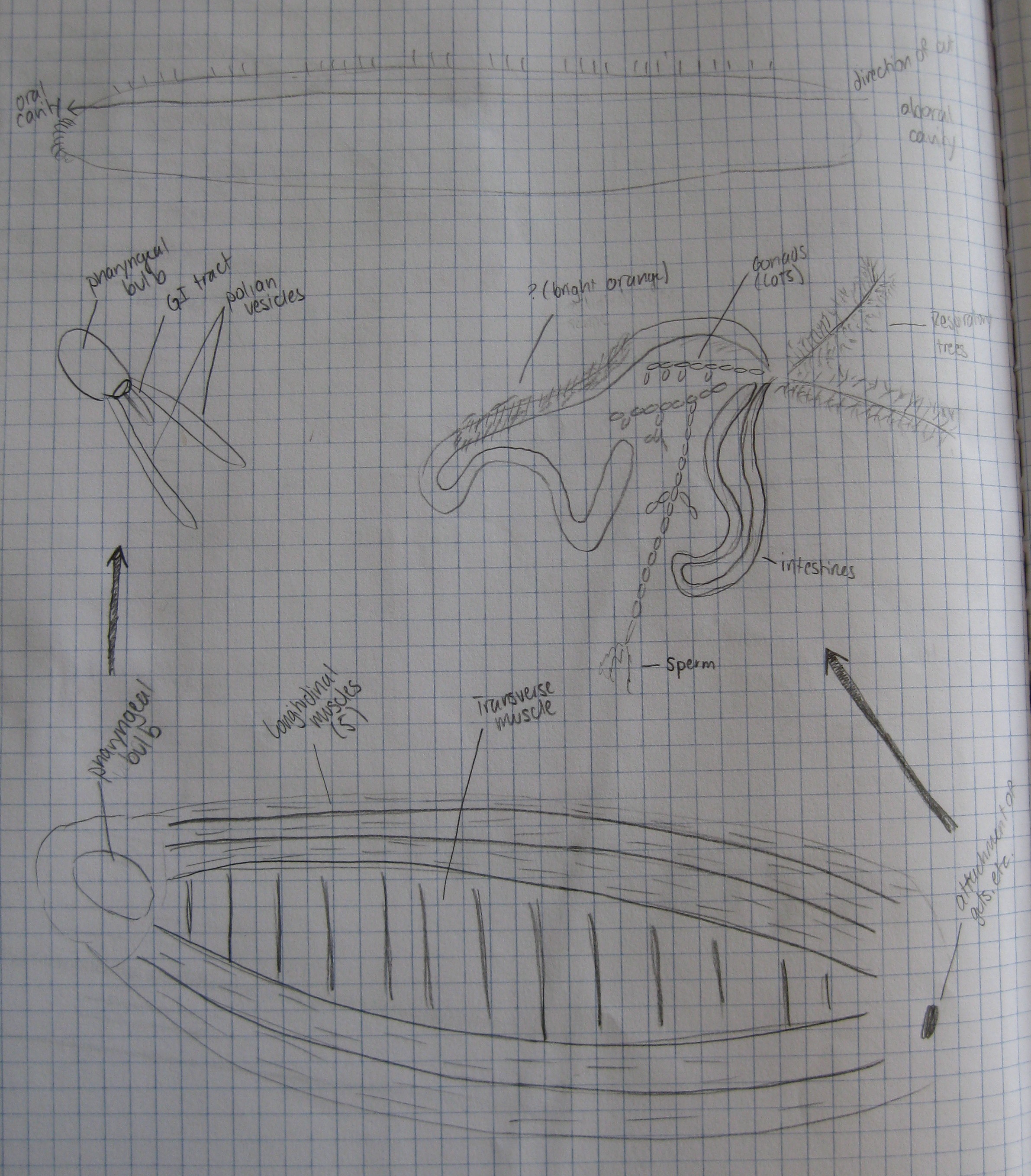
Right-hand text reads, from top to bottom:
Direction of cut
Aboral cavity
Respiration trees
Attachment of guts, etc.
I also observed the dissection of the second sea cucumber, which had a larger mass of respiratory trees and a much lower mass of gonads than the one Ashton and I dissected. I examined the results of dissections of sea stars and a sea urchin. The sea urchin, in particular, was surprisingly empty, with gonads, Aristotle's lantern, and digestive ceca all located along the internal sides of the test.
Histological comparisons:
Using pre-made slides, we compared normal and diseased tissues for oyster, abalone, and Armina under a compound microscope.
Some diagrams: